Abstract
Chronic myelomonocytic leukemia (CMML) is an aggressive myelodysplastic/myeloproliferative neoplasm represented by abnormal, clonal monocytosis, resistance to conventional chemotherapy, and an elevated risk of transforming into acute leukemia. While allogeneic hematopoietic stem cell transplantation is the only curative treatment option, this is not available to everyone due to either lack of compatible donors or the existence of comorbidities that make the patient ineligible for such procedure. Hypomethylating agents (HMAs) such as Decitabine and Azacitidine have shown efficacy in ∼50% of the patients. However, response to these agents can take 6 months or longer and patients are required to stay on the drug for this period of time, even if only about 50% of them will benefit from it. Therefore, in order to provide the best treatment option at an appropriate timing, robust biomarkers for predicting the response to HMAs are urgently needed. Using genome-wide methylation analysis (ERRBS: enhanced reduced representation bisulfite sequencing), we previously identified 21 epigenetic biomarkers which accurately predicted the response to decitabine in patients with CMML (Meldi K, et al. J Clin Invest. 2015). However, due to the complexity of this assay as well as the genome-wide nature of it, ERRBS is not suitable for the clinical setting. Thus, we sought to develop a novel, targeted assay that can be used in the clinical setting for the implementation of this biomarker as well as to validate the performance of the epigenetic biomarker in an independent cohort of patients.
For this purpose, primers targeting the 21 regions included in the epigenetic biomarker were designed with overhang adapter sequences compatible with illumina multi-index system. Genomic DNA was extracted from CD34+ human blood cells and bisulfite conversion was performed using EZ DNA Methylation Kit (ZYMO Research). Multiplex PCR parameters were established by calculating the Gibbs Free Energy (dG) for each primer combination using PrimerSuite program. Four primer pools were determined based on the dG and optimum annealing temperatures at single PCR. Multiplex PCR was performed using FastStart High Fidelity PCR System (Roche) and the combined PCR amplicons were purified by AMPure XP (Beckman Coulter). Purified amplicons were indexed by Nextera XT index kit (illumina) and KAPA HiFi HotStart ReadyMix (KAPA Biosystems) and sequenced on an Illumina MiSeq sequencer (2x150 bp). Sequencing reads were mapped to human reference genome (hg19) using Bowtie2, and cytosine methylation states were determined by Bismark.
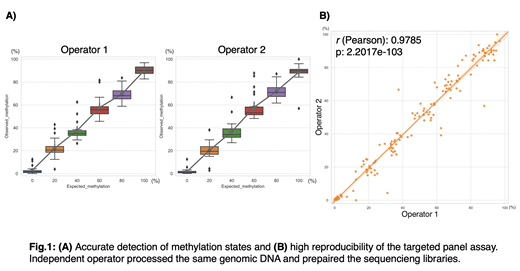
Twenty out of 21 biomarkers were successfully amplified and sequenced. Using a standard curve generated with in vitro methylated DNA, we demonstrated that the assay accurately captured the different methylation percentages in the standard curve and was highly reproducible, with correlation coefficient r=0.98 between two independent operators (Fig.1). Moreover, analysis of 26 CMML patient samples previously analyzed by ERRBS showed strong correlation between ERRBS and the targeted panel assay (Pearson's r: 0.83-0.98). To test the performance of predicting decitabine response, we analyzed both the ERRBS and the targeted panel assay data of the 26 patients (12 responders, 14 non-responders) using supervised machine learning algorithm, SVM classifier. The two assays showed 100% concordance of predictions with an accuracy of 96%, indicating high reproducibility and predictive performance of the novel targeted panel assay.
In summary, we have developed a fast, robust, and highly reproducible MiSeq-based, targeted panel assay for decitabine response prediction in patients with CMML.
Santini: Astex: Membership on an entity's Board of Directors or advisory committees; Takeda: Membership on an entity's Board of Directors or advisory committees; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees; Menarini: Membership on an entity's Board of Directors or advisory committees; Gilead: Membership on an entity's Board of Directors or advisory committees; Geron: Membership on an entity's Board of Directors or advisory committees; BMS/Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal